- Plenty of Room
- Posts
- Sidewinder Rewrites DNA Assembly Rules With Designed DNA Junctions!
Sidewinder Rewrites DNA Assembly Rules With Designed DNA Junctions!
A new assembly method builds complex DNA without sequence or length constraints
Today, we have a paper revolutionizing what you learnt in your molecular biology class: DNA assembly!
Putting long DNA together has always been a problem. Now, researchers bring you Sidewinder: a new DNA assembly method that promises to solve it! Can this meeting of DNA nanotech and DNA assembly work? Read to find out!
I’m excited for this one!
Don’t keep this newsletter a secret: Forward it to a friend today!
Was this email forwarded to you? Subscribe here!
Rewriting DNA Assembly
Researchers introduce Sidewinder, a new DNA assembly method that decouples assembly instructions from the final DNA sequence, enabling robust, high-fidelity production of long and complex DNA. Image credit: Nature.
DNA encodes the basic information of all life.
Understanding what’s written in DNA has pushed biology and biotechnology to new levels. Genetic screenings for diseases, sustainable chemical production, and personalized medicine.
All of it rests on the information → biological function relationship.
At the base of this all? The ability to read, edit, and write genetic information.
Reading and Editing: Incredible Progress
DNA reading is common now.
With next-generation sequencing platforms like Illumina, Nanopore, or PacBio, you can generate terabytes (!) of data from a single experiment, for a price that would have looked absurd just 10 years ago.
DNA editing, you ask?
CRISPR-Cas systems revolutionized DNA editing over the last decade, thanks to their ease of use and versatility. Now, you have tens, maybe hundreds of variants to choose from! For all your editing needs.
Writing DNA: Not So Great
But writing DNA? We’re way behind there.
DNA writing means assembling genetic sequences from scratch, often new genes. Ideally, we’d be able to construct DNA of any length, complexity, and diversity.
In reality, we’re nowhere close.
We’re great at synthesizing short DNA oligos (<150-200 nt). Beyond that, traditional chemical synthesis loses efficacy and accuracy. And genes can be thousands of nucleotides long.
There are alternatives. Some companies use enzymatic synthesis to create longer and more complex sequences. Promising, but it doesn’t solve the problem. For now, scientists still take short DNA fragments and assemble them into genes.
The Assembly Bottleneck
There are many assembly methods.
Nearly all rely on complementary single-stranded DNA overhangs to guide the assembly. Two matching overhangs bind, form a double helix at a two-way junction, and enzymes seal the nick. You got your strand, and those overhangs are part of the final DNA.
This works with two pieces, but what if you want to string together 5 pieces into a 1,000-nt gene? Now everything depends on the overhangs being perfectly specific.
This creates a problem: the overhangs guide the assembly, but are also part of the final product. You can’t optimize one without compromising the other! The sequences are constrained by what you’re trying to study.
This creates more than frustrated scientists. It brings real limits in:
Size: Long sequences, like genes, are hard to build and are often avoided.
Complexity: High content of guanine and cytosine (GC) and repetitive sequences break synthesis and assembly.
Efficiency: Assemblies fail, mis-ligate, or produce many side-products.
Sidewinder: A Step Up in DNA Assembly
Okay, pretty bleak picture.
But DNA assembly is painful. You spend lots of time, and you never know if it’s going to work. So, how do we fix it? Maybe by copying some DNA nanotech…
That’s exactly what the authors of today’s paper did!
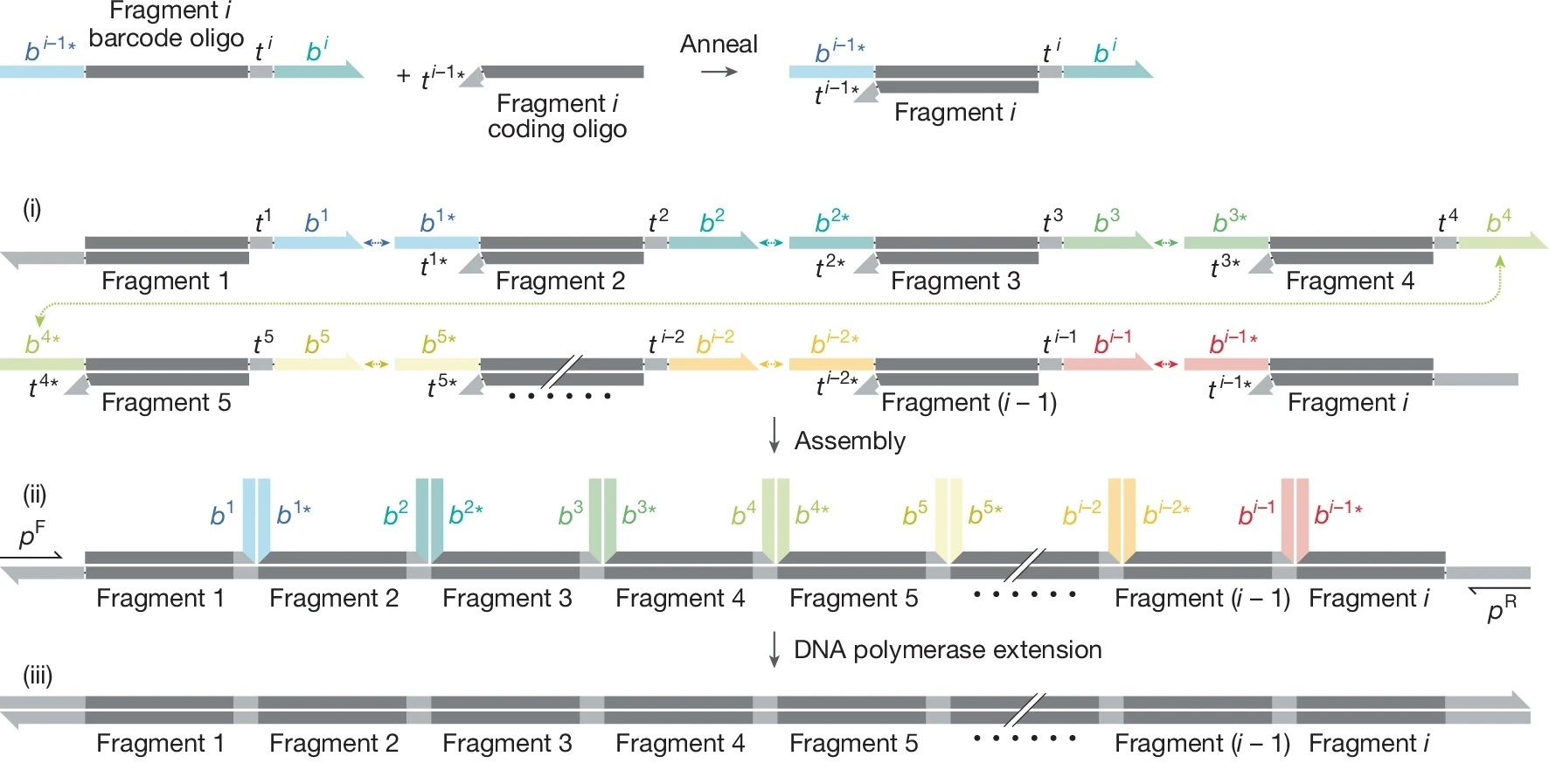
They invented Sidewinder, a DNA assembly method that uses three-way junctions (3WJs) to decouple assembly instructions from the final DNA sequence. The assembly instructions are encoded in “barcode” sequences that never end up in the product.
Because they are temporary, these barcodes can be heavily optimized, giving you robust, high-fidelity assembly without constraints in length or sequence complexity.
How Sidewinder Works: 3-Way Junctions to the Rescue
Three-way junctions are non-canonical DNA structures used in DNA nanotech.
Instead of two helices meeting, three double-helical arms converge at a single point. That third arm (the Sidewinder helix) guides the assembly process and is then removed afterward.
This eliminates constraints on where assembly happens, what sequences are assembled, and how many fragments are joined at once!
Each assembly fragment is a preformed duplex made from two oligos:
A coding oligo: Provides the final DNA.
A barcode oligo: Carries the Sidewinder barcode sequences.
Each fragment exposes two short complementary single-stranded toeholds that are part of the final construct, and two longer single-stranded Sidewinder barcodes.
Assembly happens in four steps:
Barcodes bind first
At a temperature where the toeholds are unstable, the long barcodes pair, bringing the correct fragments into a 3WJ.Toeholds engage
Inside the 3WJ, the short toeholds and pair, leaving a nick.Ligation
The nick is sealed, and the fragments are connected!Barcode Removal
The Sidewinder helix is displaced during a PCR or enzymatically removed later.
The final product is a scarless, double-stranded DNA!
Experimental Proof: Longer, Harder, and More
Once they showed the simple case, the team went big.
Literally.
Large multifragment assemblies
They assembled a gene segment from 5, 10, 20, and 40 (!) pieces. Sidewinder produced a single, strong, and correctly sized product even at 40 pieces! In comparison, traditional methods (PCA, Gibson, Golden Gate) failed beyond 5-10 fragments.
Nanopore sequencing validated the assembly. Out of the ~600 reads analyzed, over 96% were products, and 100% of the products were correct 40-piece constructs!
Analysing the junctions, they found over 22,500 ligated junctions. Error? Zero!
Hard sequences: GC-rich and repetitive DNA
Some DNA sequences are hard to assemble:
High GC content: Secondary structures can make PCR fail
Highly repetitive: PCR can fail, and toehold annealing is unreliable.
The perfect stress test for Sidenwinder.
The team tested two genes:
APOE: This cholesterol-regulating gene has 70% GC content, with peaks of 95%! A 12-piece Sidewinder assembly produced a single clean product. The sequencing showed 50,636 correctly ligated junctions out of 50,636 (!).
H-fibroin: This highly repetitive gene encodes a silk protein with promising biomaterial applications. Sidewinder produced a 500-bp DNA with a single side product and 99.77% junction accuracy.
The conventional methods? They failed.
Parallel assemblies and combinatorial libraries
Could Sidewinder do more than one thing at once?
The team mixed fragments for 3 different 10-piece constructions (mScarelt, mGL, aeBlue) in a single tube. They recovered each construct (or the whole pool), and bacterial colonies lit up after transformation!
They then went further and tested combinatorial libraries. These are collections of genetic variants with mutations in multiple positions, used in high-throughput screenings and protein design.
The researchers built a combinatorial eGFP library, with:
10 fragments
17 diversified positions
Over 440,000 variants!
Sequencing revealed 35 million junctions, with only 37 errors!
And the team recovered variants with blue/green/yellow/red fluorescence, showing functional diversity.
Sidewinder: A New Era for Biology?
An incredible work!
So much of modern biology exists because we can clone DNA, but we were always limited by our ability to make high-quality DNA. For years, it felt like the most interesting biology was hiding behind DNA that was just too hard to assemble.
Sidewinder might solve that!
There are still a couple of things to iron out. Some steps are hands-on, and the assemblies so far reached up to ~ 1 kb. Respectable, but not groundbreaking.
But I’m sure the team has more up their sleeves, since they already spun out a company. Exciting! Such big potential.
But don’t just trust me: go, read the paper here, and draw your own conclusions!
If you made it this far, thank you! What do you think of Sidewinder? Do you think it will help you in the lab? Are you going to try it? Reply and let me know!
P.S: Know someone interested in SynBio? Share it with them!
What did you think of today's newsletter?Your feedback helps create the best newsletter possible! |
More Room:
DNA Bridges for Artificial Cells: Artificial cells are the Holy Grail of synthetic biology. At this point, they can kinda mimic cellular functions, but interacting with living mammalian cells remains difficult. In this study, the authors introduce DNA-based stimulable artificial cells (STARMs) that communicate with mammalian cells through contact-dependent signaling. Built by DNA self-assembly and phase separation, STARMs contain DNA-based cytoplasm and membrane compartments that integrate functional nucleic acids and light-responsive elements. These artificial cells can sense external stimuli and trigger programmed cellular responses, including tissue formation and signaling, and show light-guided therapeutic activity in vivo, highlighting their potential for regenerative medicine.
DNA Origami Springs into Action: If you want to build a nanorobot, DNA origami is your friend. But they are not without limitations. For example, DNA origami nanorobots typically rely on simple two-state switches, limiting them to single stimulus–response behaviors. This study introduces reconfigurable DNA origami arrays that couple multiple two-state systems into a programmable network. This design allows nanorobots to process multiple inputs, perform multilevel Boolean logic, and execute complex operations with controlled timing and spatial organization. Advanced DNA origami!
AI’s Fight Against the Flu: I got the flu recently, and it wasn’t fun. I didn’t get the vaccine, but every year it’s a gamble if it will work. Now, AI is joining the fight. In this study, the authors introduce VaxSeer, an in silico framework that predicts how well vaccine strains will match future circulating viruses by combining sequence, antigenicity, and dominance data. Tested on 10 years of retrospective influenza data, VaxSeer consistently outperforms annual strain recommendations and shows a strong correlation with real-world vaccine effectiveness and disease burden. The work highlights how computational prediction can improve vaccine strain selection.
Share Plenty of Room with founders or builders
I help biotech and deep tech companies transform complex technologies into engaging content that builds credibility with investors, partners, and potential hires. Let’s chat!
Know someone who’d love this?
Pass it on! Sharing is the easiest way to support the newsletter and spark new ideas in your circle.Got a tip, paper, or topic you want me to cover?
I’d love to hear from you! Just reply to this email or reach out on social.