- Plenty of Room
- Posts
- AI-Designed Enzyme Atoms: RFdiffusion2 Crafts Catalytic Proteins!
AI-Designed Enzyme Atoms: RFdiffusion2 Crafts Catalytic Proteins!
Designing functional enzymes by placing catalytic atoms with RFdiffusion2
Today I’m a bit sick, it’s flu season!
But that didn’t stop me from bringing you a cool paper! Computational enzyme design is all the rage these days, and RFdiffusion is at the center of it.
But can it be made even better? Read on to find out!
Don’t keep this newsletter a secret: Forward it to a friend today!
Was this email forwarded to you? Subscribe here!
Enzymes from Atoms
Scientists introduce RFdiffusion2, a generative AI model that can create enzymes from scratch with atomic precision. Image credits: Nature
Enzymes: Engines of Life
You all know that enzymes are the workhorses of life.
They accelerate chemistry, turning reactions that would take a lifetime into a matter of nanoseconds. This puts them firmly at the center of the biotech revolution!
The DNA polymerase that powers PCR and molecular cloning, CRISPR-Cas systems that save lives via personalized gene editing, and the enzyme that makes your laundry detergent more efficient and sustainable!
The industrial applications of enzymes are endless.
But finding natural enzymes for new reactions? That sucks. You look at countless proteins in databases, pick one, do high-throughput screenings, and hope something works.
Not great!
Computational protein design promises a better way. Today, you can create DNA and protein binders, or molecular LEGO blocks, with nothing but your laptop (and some experiments later)!
But designing an enzyme from scratch is an incredibly hard task!
De Novo Enzymes: The Design Holy Grail
De novo enzyme design aims to generate enzymes that catalyze new reactions. That’s the dream!
You can think of an enzyme as two parts:
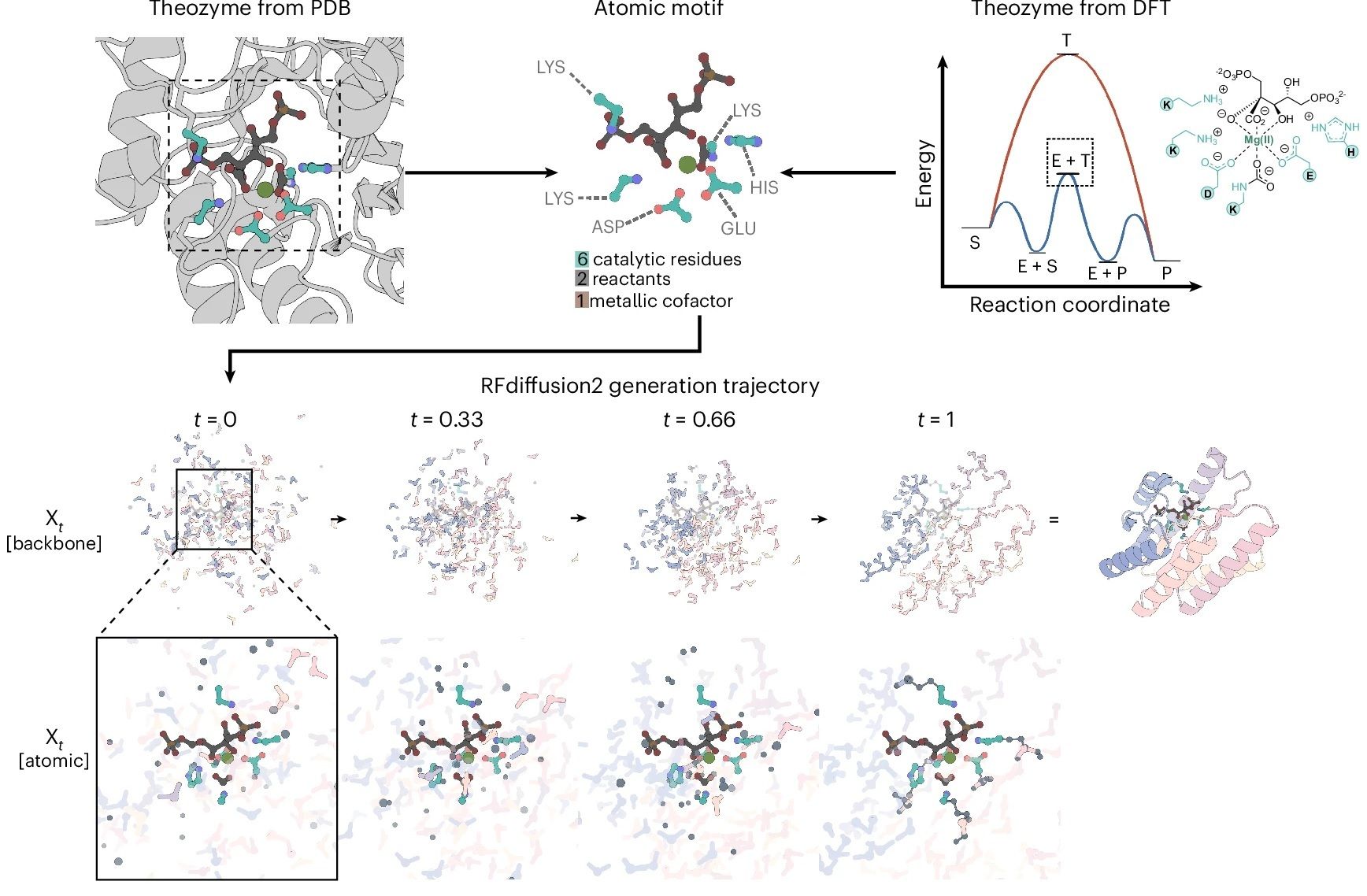
Active site (the theozyme): the catalytic residues that make the reaction happen.
Protein scaffold: The rest of the protein that keeps the active site in the right geometry and creates the right environment.
Protein design in general, and enzyme design in particular, has seen a revolution with the arrival of deep learning models.
Using diffusion models, is now possible to sample many different protein scaffolds, custom-built around your catalytic site! This process, known as motif scaffolding, ensures a much higher degree of success.
But these systems can still be improved.
In most approaches, proteins are represented at a “backbone-level” and treated as a sequence of amino acid residues and a chain of backbone atoms. This ignores the side chains, where the magical chemistry happens!
This is a real limitation! Enzymes work because the functional groups in the side chains are positioned with extreme precision. Even an error of <1 Å can destroy activity.
Scientists overcome this by brute force. They create millions of possible positions for the catalytic residue and then filter them to keep the most plausible ones.
This works, but it’s computationally intense. And with more complex active sites, it’s practically impossible!
Upgrading Diffusion Models: RFdiffusion2
If that sounds familiar, you’re paying attention.
Just a couple of weeks ago, we saw how scientists built exactly one of these “brute force” approaches to enzyme design. And the pipeline they created worked!
But can we get better tools and avoid this altogether?
Today’s paper brings us RFdiffusion2. RFdiffusion is a generative model that creates proteins from scratch:
Initialization: It starts with a random "cloud" of amino acid residues, essentially noise with no structure.
Progressive Denoising: Over many iterations, the noise resolves into a well-folded protein structure tailored to the target.
Output: The final product is a designed protein that matches the desired shape and properties.
This system has become a staple tool in the protein design toolbox, but it doesn’t have atomic-level precision.
And that’s exactly where RFdiffusion2 brings an upgrade. It is trained on atom-level active site descriptions, and it can simultaneously predict where a residue should be (the residue indices) and its orientation (the rotamers)!
And all this while generating a protein scaffold to accommodate the atoms of the active site. No enormous screenings of all possible positions!
Training and Pipeline: What’s New Under the Hood
I’m not the most qualified person to talk about this, so I’ll just make a highlight reel.
During training, some residues are represented as fully-atomized residues, with the atoms of the side chains also provided. In this way, the model learns to place motif atoms into the best positions during generation.
In addition, some residues are provided “unindexed”, so the model learns where to place them. This enables you to work with theozymes where catalytic residues don’t have known indices!
A cool detail: the model was trained for 17 days on 24 A100 GPUs! I wonder how much that cost… The training dataset was mostly composed of proteins, protein-small molecule, and protein-metal complexes from PDB!
With this new training, the pipeline looks simple:
You input an active site, as residues or as atomic coordinates!
RFdiffusion2 creates a protein scaffold around it.
LigandMPNN (another deep learning tool) designs sequences from 3D protein structures.
Chai-1 (an open source tool) predicts the structure from the designed sequences.
The predicted structures are scored against the designs. A design will pass the filter if the active site in the predicted structure is within 1.5 Å of the target motif and if there are no ligand clashes.
Voilà!
This pipeline was used for the enzyme design campaigns, but also to benchmark RFdiffusion2. The authors showed that it outperforms its predecessor, RFdiffusion, especially with more complex motifs!
Experiments: 3 (+1) Enzyme Campaigns!
The authors tested RFdiffusion2 experimentally in 2 ways:
Scaffold minimal active sites from native enzyme structures.
Design from first principles by deriving transition-state geometries from modeling.
The team screened ≤96 designs per reaction and found active enzymes in every case! Pretty incredible.
Let’s see what they worked on!
Retro-aldolase campaign
Retro-aldolases break carbon-carbon bonds in molecules and are a well-studied enzymatic system.
The authors screened 96 designs in an in vitro transcription/translation assay. Four variants had detectable activity, with the best design having a kcat/KM ≈ 6.34 ± 0.92 M⁻¹ s⁻¹. Not incredible, but it’s catalytic acceleration!
A couple of weeks ago, I covered Riff-Diff, a different pipeline for enzyme design based on RFdiffusion. The authors there also studied retro-aldolases! And their best design had a kcat/KM of 290 M⁻¹ s⁻¹.
The two teams started with the same system, and reached pretty different catalytic rates! For reference, most enzymes have a kcat/KM of around 105 s−1M−1. So, good starting points to optimize!
Cysteine hydrolase campaign
Cystein hydrolases degrade proteins and are used in many industrial applications.
The team checked 48 designs, finding several with activities. The best one had a kcat/KM ≈ 248 ± 34 M⁻¹ s⁻¹! Much better than the previously designed cystein hydrolases.
Zinc metallohydrolase
Last but definitely not least, the coolest campaigns.
Metallohydrolases coordinate metal ions and use them to hydrolyze molecules. If we could design new enzymes that use this mechanism, we could destroy, for example, durable pollutants!
The researchers used modeled active sites as a starting point to design enzymes for two substrates, testing 96 designs per substrate:
4MU-butyrate: The best kcat/KM was around 77 ± 10 M⁻¹ s⁻¹, better than previous designed zinc hydrolases
4MU-phenylacetate: The best design had a crazy kcat/KM of 16,000 M⁻¹ s⁻¹! This is orders of magnitude better than previous designs. And in a second campaign, they found a design with kcat/KM 53,000 ± 5,000 M⁻¹ s⁻¹! Awesome.
RFdiffusion2: A Worthy Sequel
Well, this was cool!
It’s amazing that we’re coming closer to useful de novo designed enzymes. The opportunities are limitless! Enzymes fuel life, after all. We could solve many problems, from diseases to pollution (think about all that plastic…).
RFdiffusion became a staple of the protein design world, fast. Will RFdiffusion2 do the same? It works even better! Maybe we’re getting closer to real, functional custom enzymes!
This was a good read! Go here to get all the details.
If you made it this far, thank you! What do you think of today’s issue? Reply and let me know!
P.S: Know someone interested in ML-based protein design? Share it with them!
What did you think of today's newsletter?Your feedback helps create the best newsletter possible! |
More Room:
Computing with DNA Origami: One day, your laptop will run on DNA origami. Maybe. In the meantime, in this study, the authors introduce a combinatorial rule-based strategy using 3-input, 1-output logic rules to generate a wide variety of patterns with fewer unique DNA tiles. DNA-based algorithmic self-assembly enables nanoscale computation using logic encoded in DNA tiles, but scaling to complex patterns typically requires many unique components. By combining complementary and non-complementary rule sets, they expand the range of computable functions while improving assembly efficiency. Atomic force microscopy confirms that the resulting DNA lattices closely match theoretical predictions.
Synthetic Mechanoreceptor Stimulation: Funny story: the original project for my PhD was supposed to use DNA origami to activate mechanoreceptors in cells. That never really worked out, but I’ve kept a soft spot for the subject. This review summarizes recent advances in engineering synthetic mechanoreceptors, covering both genetic approaches that reprogram natural receptors and non-genetic strategies based on DNA nanotechnology. DNA-based mechanosensitive devices enable programmable control over receptor organization and force responsiveness, including DNA-functionalized artificial mechanoreceptors that impart mechanosensitivity to otherwise non-mechanosensitive receptors without genetic modification. Together, these advances position DNA nanotechnology as a powerful toolkit for studying mechanobiology and developing force-guided therapeutic strategies.
Unfolding RNA Hybrids: Sometimes, common chemicals can really help in the lab. DNA and RNA nanostructures can be tuned through interactions with small molecules, but simple and scalable methods are limited. In this study, the authors show that urea can modulate the mechanical stiffness of RNA:DNA hybrid nanostructures through supramolecular interactions with the helix. Using nanopore sensing and atomic force microscopy, they demonstrate that urea-stiffened hybrids exhibit fewer folded translocations, improving signal quality. This approach enhances single-molecule nanopore detection and supports more robust sensing of low-abundance RNA targets.
Share Plenty of Room with founders or builders
I help biotech and deep tech companies transform complex technologies into engaging content that builds credibility with investors, partners, and potential hires. Let’s chat!
Know someone who’d love this?
Pass it on! Sharing is the easiest way to support the newsletter and spark new ideas in your circle.Got a tip, paper, or topic you want me to cover?
I’d love to hear from you! Just reply to this email or reach out on social.