What if you could use DNA to store your data, and then use CRISPR-Cas9 to get it out? Well, this is exactly what we’re exploring today! Yes, there is more than gene editing to CRISPR-Cas9.
Also, another big milestone. Today marks one year of Plenty of Room! 🎂🎉 Thank you for being here. It has been a fun ride!
And I’m not stopping anytime soon, don’t worry 😎
CRISPR-Based Google
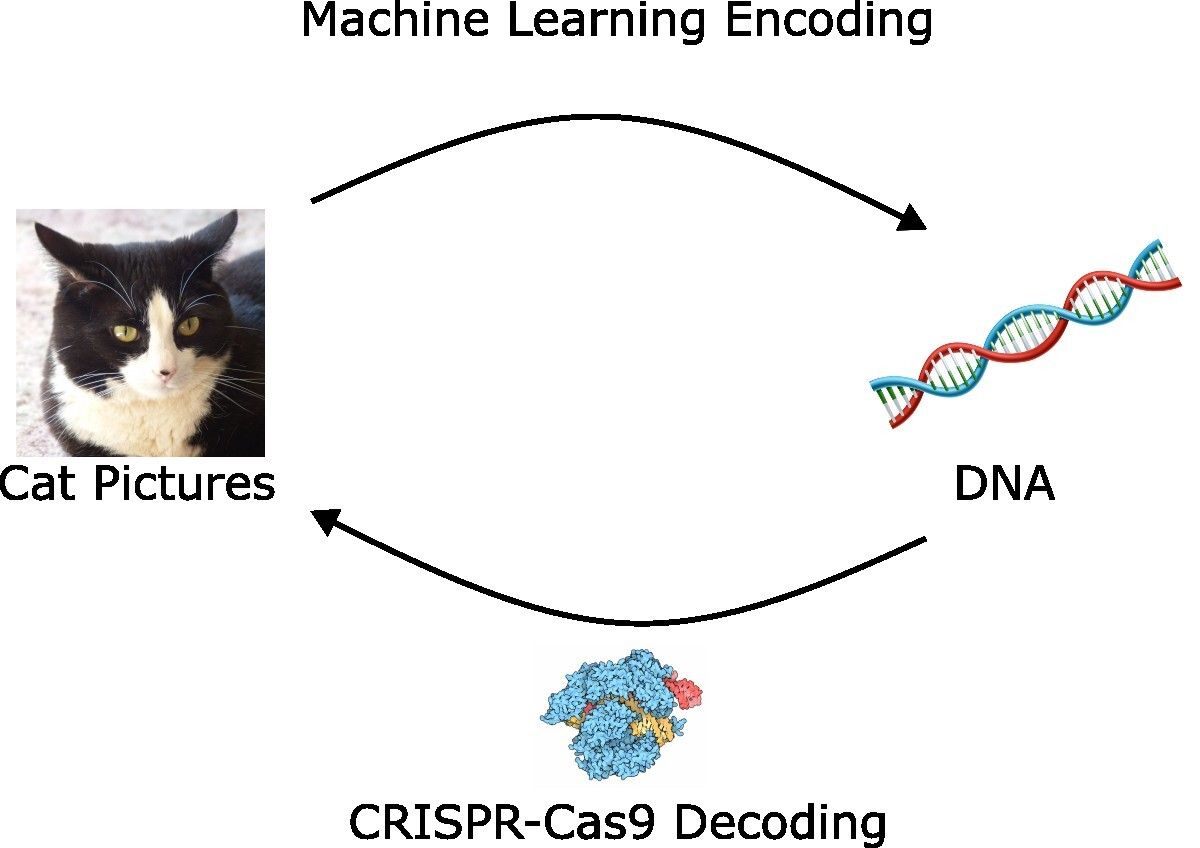
Scientists used machine learning to encode data into DNA and decode it using CRISPR-Cas9. And yes, they used cat pictures.
Drowning in Data
Do you know what we produce a lot of? Data.
From cat videos at the start of the internet, to TikTok and ChatGPT today, we create absurd amounts of data. We are talking about 400 million terabytes of data per day! And while it doesn’t feel real, all this data is physically stored in data centers around the world.
But we are running out of storage space, and data centers are energy-hungry beasts. That’s why there is a lot of research to find new materials for data storage. And between glass beads and semiconductors, there is a familiar face poking out: DNA.
DNA works great for data storage. Not surprisingly, it has been maintaining genetic information for millions of years. And now, apparently, it will also help keep your cat videos.
Why DNA Data Storage?
DNA has great characteristics for this:
Ultrahigh information density: 1 gram of DNA can store 1 million TB of data! That’s a lot.
Stability: DNA is stable for thousands, and even millions of years!
Low cost: Sequencing costs have fallen, and synthesis is following. Soon it will be a cheap alternative!
People have already created end-to-end workflows for encoding, synthesis, retrieval, sequencing, and decoding! But there are still challenges: sorry, no DNA laptop for you yet.
Data Retrieval: Where Things Break Down
One of the basic functions of a data storage system is the search and retrieval of files.
In the early days of DNA data storage, you had to sequence the whole library to retrieve a single file. Not convenient! But scientists solved this using random access, the ability to access any data directly, without needing to see other elements first. So, you can pick your file, without having to go through all the other ones!
The most common approach is to use PCR. But it has limits:
Slow and energy-intensive: Often, these protocols need long incubations in a thermocycler at high temperatures
Primer design is complicated
Poor multiplexing: Retrieving more than one file in a single reaction is challenging
These issues get worse when working with bigger databases, where DNA data storage should shine.
CRISPR-Cas to the Rescue
Here is where today’s paper comes in! The authors created a CRISPR-Cas9-based random access (C9RA) in DNA data storage. And more! But let’s not get ahead of ourselves.
They first prepared the DNA database. This is the data storage that will be accessed! This required a few steps:
Encoding of files in single-stranded DNA: They encoded 25 batches of images in DNA! Each data “file” is encoded with a unique 20-nt Cas9 target sequence.
Conversion to double-stranded DNA: Using Gibson assembly, in preparation for the next steps
Rolling circle amplification: This isothermal amplification allows them to obtain multiple copies of each strand. By the way, the method used is called rolling circle amplification to concatemeric consensus, or simply R2C2. Yeah, it’s a Star Wars reference.
Once the library is ready, it’s time for retrieval! Cas9-RNA complexes are programmed with file-specific guide RNA. This way, Cas9 cuts only the DNA strands matching the gRNA, exposing ends for adapter ligation for Nanopore sequencing.
They tested the capabilities of the system:
100x enrichment for targeted files when recovering only one file (only file 10, for example)
Multiplexing successful! Two orders of magnitude enrichment for targeted files, and they tested up to 20 files!
But the team didn’t stop here.
Similarity Search: Google Image, But in DNA
Okay, until now, their retrieval system worked if you knew exactly the sequence of the file to search.
But modern search systems don’t work like that; they use content similarity. One example is reverse image search. You search for something, and the data are retrieved based on resemblance.
And doing that with DNA is hard! So, the team created Cas9-based Semantic Search (C9SS).
For the first step, they encoded a database of 1.7 million images into DNA sequences via a neural network. This network was trained to maximize Cas9 cleavage rates between similar items: for example, all images of a cat will have similar sequences for Cas9!
For the retrieval, they encoded a query image into the Cas9 gRNA. The Cas9-gRNA complex cleaves both exact and similar sequences in the DNA pool, which are then sequenced with Nanopore!
And the results were pretty good:
The system worked to recover similar images of cats and of Bigfoot!
In the case of the cat, 98% of the similar images were retrieved!
Can I Store My Data in DNA?
We are not there yet! But we are moving closer. This new technology has great advantages:
It saves energy, since it works isothermally at 37°C
It’s fast, requiring only a few minutes, compared to hours for other systems
It allows multiplexing (so I can get all my cat pictures at the same time)
It’s compatible with automation
It’s not perfect, though:
Destructive retrieval: Cas9 cuts the target DNA
Precision: There were off-target retrievals, especially in the similarity search
But it’s possible to improve! For example, using Cas9 that binds without cutting or combining the two systems: C9SS first for broad filtering, then C9RA for precision.
This was a cool paper! I had to skip over some details, so go here and read them all!
If you made it this far, thank you! Do you see a future for DNA data storage? Are you working on it? What do you think could be the next breakthrough? Reply and let me know!
P.S: Know someone interested in data storage? Share this with them, it will make them happy!
More Room:
Oxidizing DNA Origami: I find the intersection of DNA nanotech and “normal” nanotech super cool. This study shows that dynamic DNA origami structures can template the growth of iron oxide nanomaterials while preserving their ability to change shape and self-assemble. Despite some aggregation at high concentrations, the structures retained functional overhangs, enabling cargo binding, reconfiguration, and assembly. This could be useful for robust applications of DNA origami!
Shape-Shifting DNA Origami: There are many ways to move DNA origami, but not many leverage the four-way junctions at the base of DNA nanostructures. This study explores how the design of DNA bases at four-way junctions affects the kinetics and thermodynamics of transformations in reconfigurable DNA nanoarrays. Using a uniformly sequenced domino-style DNA array, researchers demonstrated that they could modulate the structural transformation of the array by tuning the energy landscape between different junction configurations. Cool!
Bacterial Nanopores: I love structural biology papers! This study reveals that PopA, a major outer membrane protein (OMP) from the predatory bacterium Bdellovibrio bacteriovorus, forms an unusual pentameric porin-like structure. Using X-ray and cryo-EM, the researchers found that PopA creates a bowl-shaped β-barrel assembly that encloses a section of the membrane lipid bilayer. This structure, when misdirected into E. coli, disrupts membrane integrity. I wonder if it could be used to fight bacteria!
Share Plenty of Room with founders or builders
I help biotech and deep tech companies transform complex technologies into engaging content that builds credibility with investors, partners, and potential hires. Let’s chat!
Know someone who’d love this?
Pass it on! Sharing is the easiest way to support the newsletter and spark new ideas in your circle.Got a tip, paper, or topic you want me to cover?
I’d love to hear from you! Just reply to this email or reach out on social.
