Welcome to Plenty of Room!
Plenty of Room is your guide to the cutting-edge news related to molecular machines. New here? Just go ahead and subscribe! Already subscribed? Help a friend or a colleague save some time and share this with them!
Let’s get into it now.
Is AI Coming for GFP?
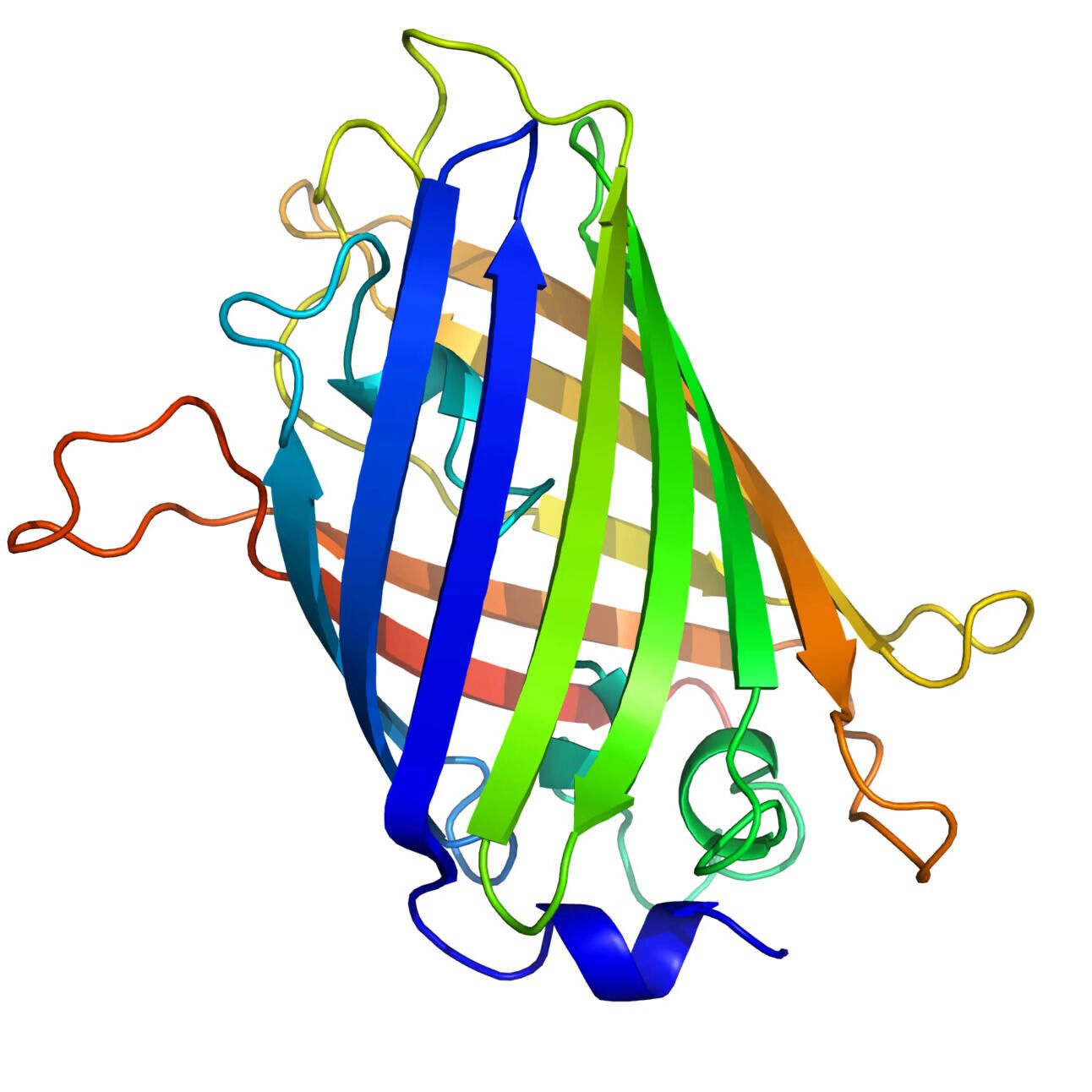
Structure of native GFP, rendered in PyMOL. Picture by Zephyris, licensed under CC BY-SA 3.0.
Who hasn’t heard of GFP? The green fluorescent protein that makes jellyfish glow has been a staple in biology for ages.
Fluorescent proteins share a simple but elegant structure: a nearly perfect cylinder formed by eleven β-strands, with a central alpha helix containing the light-emitting chromophore. Unlike other light-emitting proteins that need extra helpers, GFP creates a chromophore out of its own structure, making it a prime candidate for engineering to improve its brightness and diversify its colors. So, what better target for testing your new AI protein design model than something this iconic and easy to measure?
Introducing ESM3: AI-based Protein Design
Enter ESM3, EvolutionaryScale’s new protein language model. EvolutionaryScale is the latest entrant to apply cutting-edge machine-learning models to biology. And they made a powerful entrance, with US$142 million in funding to apply their models to sectors such as sustainability and drug development. ESM3, one of the largest language models developed for biology, was trained using the sequences, structures, and functions of over 2.7 billion proteins.
This model creates proteins based on user specifications, and the authors tasked it with generating a GFP-like protein from a few key amino acids. They synthesized and tested the 88 most promising proteins, and although most failed, one glowed faintly: the team asked the model to refine this sequence, achieving a protein that fluoresced as strongly as native GFP. The most interesting part is that the new protein’s predicted structure is similar to natural fluorescent proteins, but the sequence is very different, matching less than 60% to the closest relative: in the preprint, the authors highlight that this level of difference is equal to “over 500 million years of evolution”.
The Future of AI in Protein Design
EvolutionaryScale is stepping onto an increasingly crowded stage for generative AI in biology, with other players like 310AI, Cradle, or Profluent. I find the prospect of generating new proteins extremely exciting, and I can’t wait to see what is going to be created next with these powerful tools.
Read the preprint here.
In Other News:
Barcoding antibodies to track proteins: Mapping protein-protein interactions is hard, but this study might make it a little bit easier. The team introduced a DNA-barcoding method for multiplexed mapping of protein interactions into the cells, and they used it to link higher-order protein interactions to cancer aggressiveness.
Bacterial editing goes direct: Research is showing more and more just how important the microbiome is, but tools for editing bacterial targets are lacking. Until today: here, researchers engineered a phage-derived particle to deliver a base editor to modify E. coli directly in the mouse gut, opening the way for new microbiome-targeted therapies.
Algae wheeling away: Microalgae and micromachines: name a more iconic duo. This must have been what the scientists behind this research thought: they created microtraps to harness the propulsion power of microalgae. They even made two designs: a “Rotator” and a “Scooter”. It’s amazing, go read for yourself.
Not yet a subscriber to Plenty of Room? Sign up today — it’s free!
You think a friend or a colleague might enjoy reading this? Don’t hesitate to share it with them!
Have a tip or story idea you want to share? Email me — I’d love to hear from you!
You have something you would love me to cover? Just reach out here or on my social!
